Eric C. Anderson is a statistical geneticist focused on the application of molecular technologies to ecological research and fisheries management. He has worked extensively in the application of single nucleotide polymorphism (SNP) markers to Pacific salmon fishery management and research. Recently, with his colleagues in the Molecular Ecology and Genetic Analysis (MEGA) team at the Southwest Fisheries Science Center, he has been promoting the development of novel, next-generation sequencing methods for conservation genetics, implementing bioinformatic and statistical approaches to make the methods broadly applicable. He also contributes to the Bird Genoscape Project, which his wife co-directs, and collaborates widely with molecular ecologists, conservation geneticists, and fishery biologists.
Eric is an active proponent of reproducible research and open science. You can find that many of his ongoing research projects are developed in the open, publicly, on GitHub. He has also taught courses (in 2014 and 2017) in reproducible research using R, RStudio, and GitHub to his colleagues at NMFS, and to students and affiliated researchers at the University of California, Santa Cruz.
You can get Eric’s CV here in pdf.
PhD in Quantiative Ecology and Resource Management, 2001
University of Washington
MS in Fisheries, 1998
University of Washington
BA in Human Biology, 1993
Stanford University
Mon, May 1, 2017, Southwest Fisheries Science Center Ignite Talk
Tue, Feb 21, 2017, USC Marine and Environmental Biology, Departmental Seminar
Mon, Nov 14, 2016, Durham University Ecology Seminar

Still working on inserting info about the boatload of projects we are cooking up…

Studying larval recuitment of kelp rockfish
Mapping the flyways of America
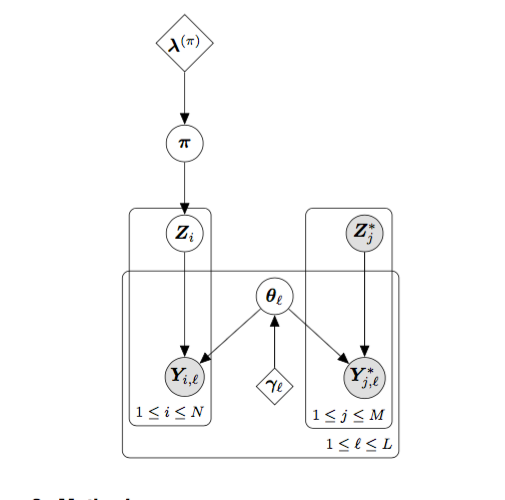
Computationally efficient genetic stock identification in the tidyverse.
Though my main appointment is at the National Marine Fisheries Service, I do get to teach a bit. Lately I have been teaching full-length courses or seminars in reproducible research methods and data analysis to colleagues and ecology students. I also get to guest lecture occasionally.
Quarter-courses:
Recent Guest Lectures
Summer Institute in Statistical Genetics
Conservation Genomics Workshop, Flathead Lake Biological Station